Crosstalk
Cells communicate with their environment via signal transduction pathways. On occasion, the activation of one pathway can produce an effect downstream of another pathway, a phenomenon known as crosstalk. We say that crosstalk occurs when the activation of one pathway (e.g., the Notch pathway) elicits a transcriptional response downstream of another pathway (e.g., the TGF-beta pathway). Figure 1. Crosstalk between pathways occurs in a directed fashion, i.e., crosstalk from the Notch pathway to the TGF-beta pathway may occur using different mechanisms from crosstalk from the TGF-beta pathway to the Notch pathway.
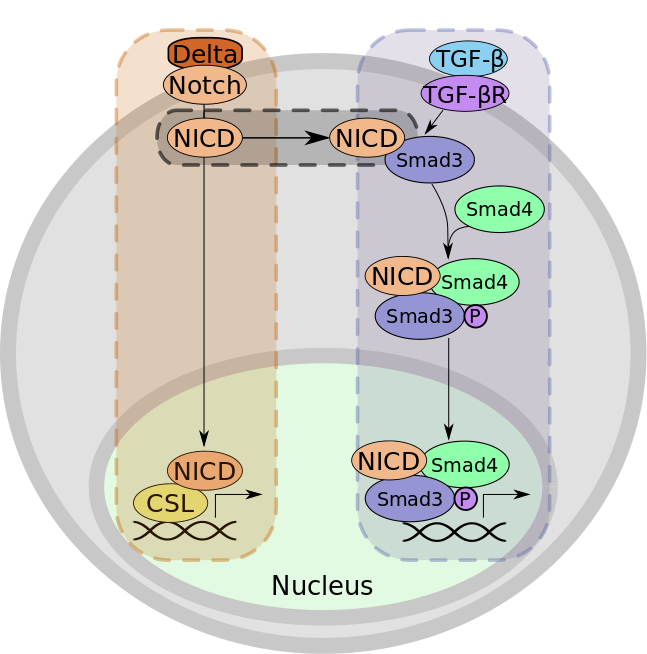
Figure 1: An example of activating crosstalk. (A) The Notch signaling pathway. When the Delta ligand binds to Notch, the Notch intracellular domain (NICD) is cleaved and translocates to the nucleus, where it binds to CSL. The NICD:CSL complex controls the transcription of developmental genes. (B) The TGF-beta signaling pathway. The binding of TGF-beta to its receptor results in the heterodimerization of SMAD3 and SMAD4 and the subsequent phosphorylation and translocation of the complex to the nucleus. Here, this complex regulates the transcription of genes involved in differentiation. (C) Crosstalk from the Notch signaling pathway to the TGF-beta signaling pathway occurs when NICD binds to SMAD3 to further promote complex formation between SMAD3 and SMAD4. After crosstalk, the transcriptional complex includes NICD (not shown in the figure).
The XTalkDB Website
XTalkDB is a carefully curated database that documents the scientific literature that supports crosstalk between pairs of pathways. For each of 650 pathway pairs, XTalkDB catalogues two types of evidence:
- We have curated literature to support the crosstalk from the first pathway to the second.
- We have yet to curate a publication that documents crosstalk between that pair of pathways.
This website provides an interface to query, visualize, and access this database. We describe its main features below:
Take a tour
On the home page you can take a tour of XTalkDB.
Search by Pathways

Figure 2: XTalkDB’s pathway search bar. The user may select one or more pathways to query the database.
A user may search XTalkDB for crosstalking pathway pairs. The search box provides a dropdown of a list of pathways contained in XTalkDB. The user may select one or more pathways from this dropdown to query the database. The results will depend on the number of pathways selected.
- One pathway: XTalkDB will return records for all pathways with which the query pathway crosstalks.
- Two pathways: XTalkDB will return all records of crosstalk between these two pathways.
- Three or more pathways: XTalkDB will return all records of crosstalk between every pair of the query pathways.
The results page when querying by pathways
TThe results page when querying by pathways includes two views: a list view and a network view.
List view
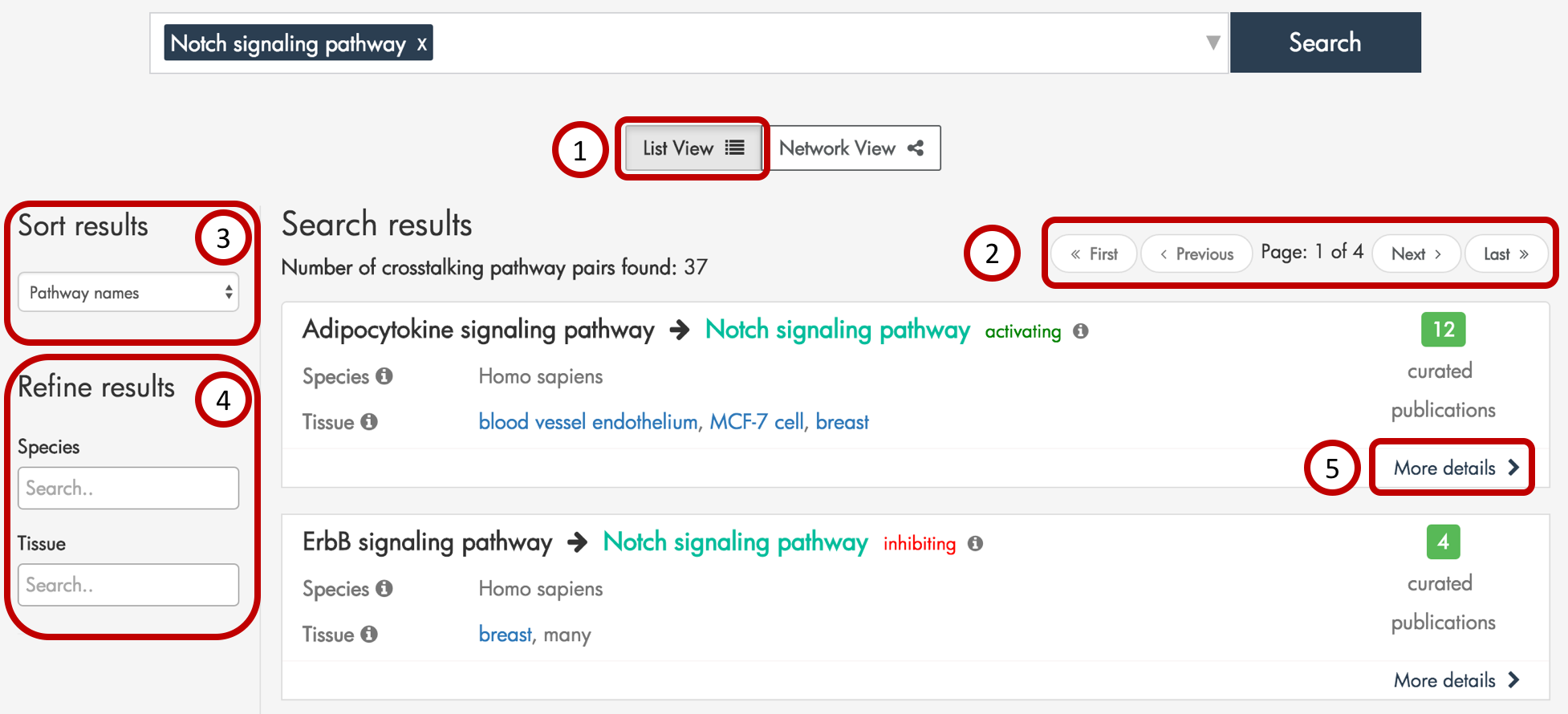
Figure 3: Example search result page when querying by pathways. Here, we display the List View.
XTalkDB displays each crosstalking pathway pair from the search results as its own item in a list (Figure 3, part 1). Up to 10 results are displayed per results page, and the user may navigate to additional pages using the ‘next’ and ‘previous’ buttons at the top (Figure 3, part 2). The user may sort the results (Figure 3, part 3) by the first pathway name or the number of publications documenting crosstalk. In addition, the user may filter the search results (Figure 3, part 4) to only display those pathway pairs documented in a particular species and/or tissue.
In the list of search results, each queried pathway is highlighted. Each result contains the direction of crosstalk, the number of publications supporting the crosstalk, the species and tissues in which the crosstalk was documented, and a link to more details (Figure 3, part 5) about the pathway pair.
Network view
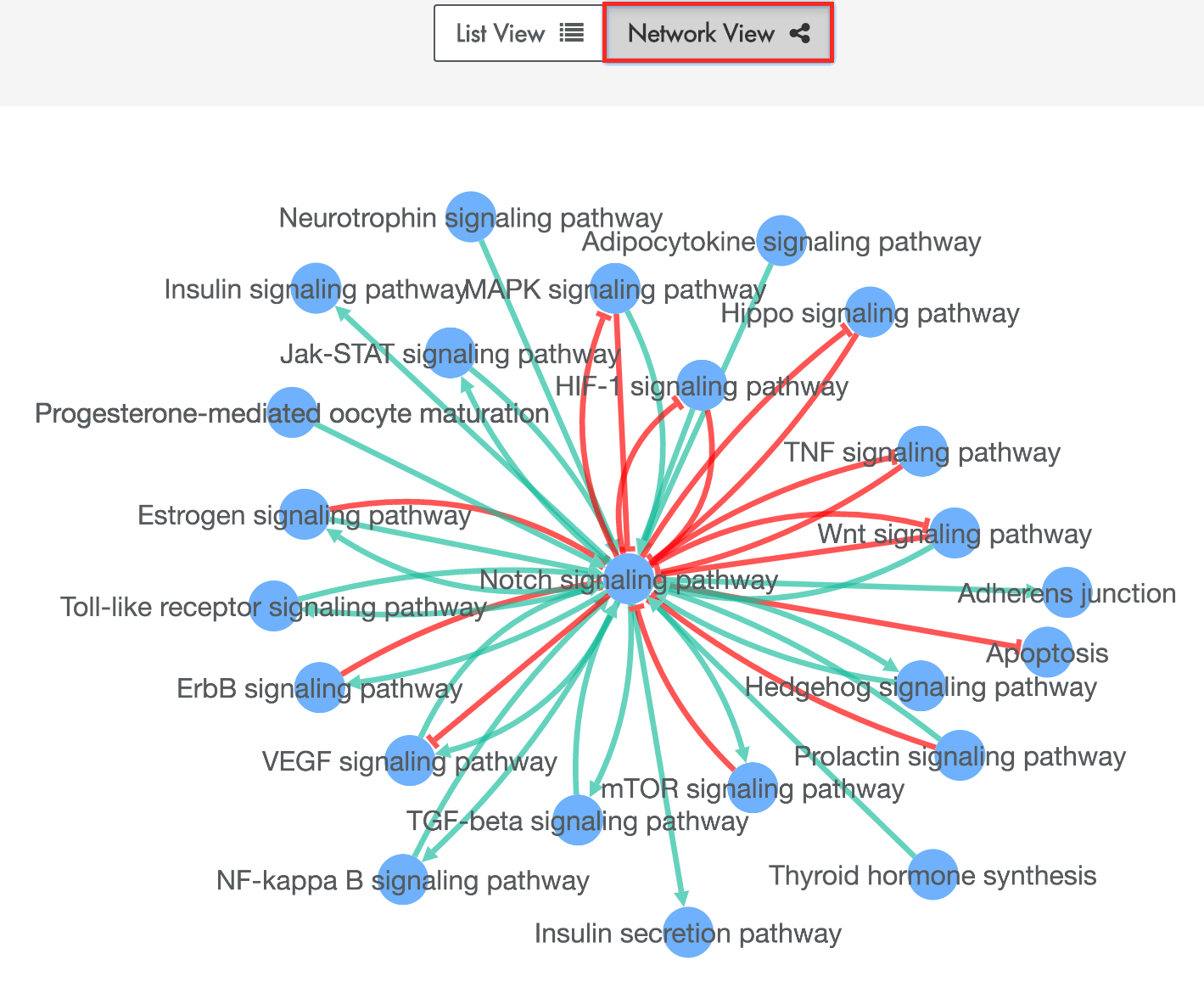
Figure 4: Example search result page when querying by pathways. Here, we display the Network View.
XTalkDB also allows a user to display the search results in a network view. The network represents the search results as a directed network of crosstalking pathway pairs. In this interactive network, each node represents a pathway and each directed edge represents crosstalk from the tail pathway to the head pathway. A user may lay out the network by selecting and dragging nodes. The user can click on an edge in this network to display a popup containing more information on the pathway pair.
Search by Molecules
A user may search XTalkDB for crosstalking pathway pairs by the mediating molecules. Figure 5. The search box provides a dropdown of a list of molecules contained in XTalkDB. The user may select one or more molecules from this dropdown to query the database. The results will depend on the number of molecules selected.
- One molecule: XTalkDB will return records for all pathway pairs with which the query molecule mediates the crosstalk.
- Two or more molecules: XTalkDB will return records for all pathway pairs with which at least one of the query molecules mediates the crosstalk.
XTalkDB uses partial string matches when querying by molecules. For example, a query for 'VEGF' will return results for both 'VEGFA', 'VEGFB' and 'VEGFC'.

Figure 5: XTalkDB’s molecule search bar. The user may select one or more molecules to query the database.
Crosstalk page
XTalkDB contains one page for each pair of crosstalking pathways. This page summarizes all the curated evidence of crosstalk contained in the database.
Curated literature with evidence for crosstalk

Figure 6: Page displaying the results for the crosstalk from Notch to TGF-beta signaling pathways. Publications with evidence for crosstalk are displayed at the top of the page.
XTalkDB lists all publications supporting the crosstalk between the two pathways. For each curated publication supporting crosstalk, XTalkDB displays the publication title, PubMed ID, molecules mediating the crosstalk (if known), species, tissue, whether the crosstalk is transcriptional, and the regulation type (i.e., activating/inhibiting). Figure 6. See Downloads section for more details on each attribute.
Molecules mediating the crosstalk
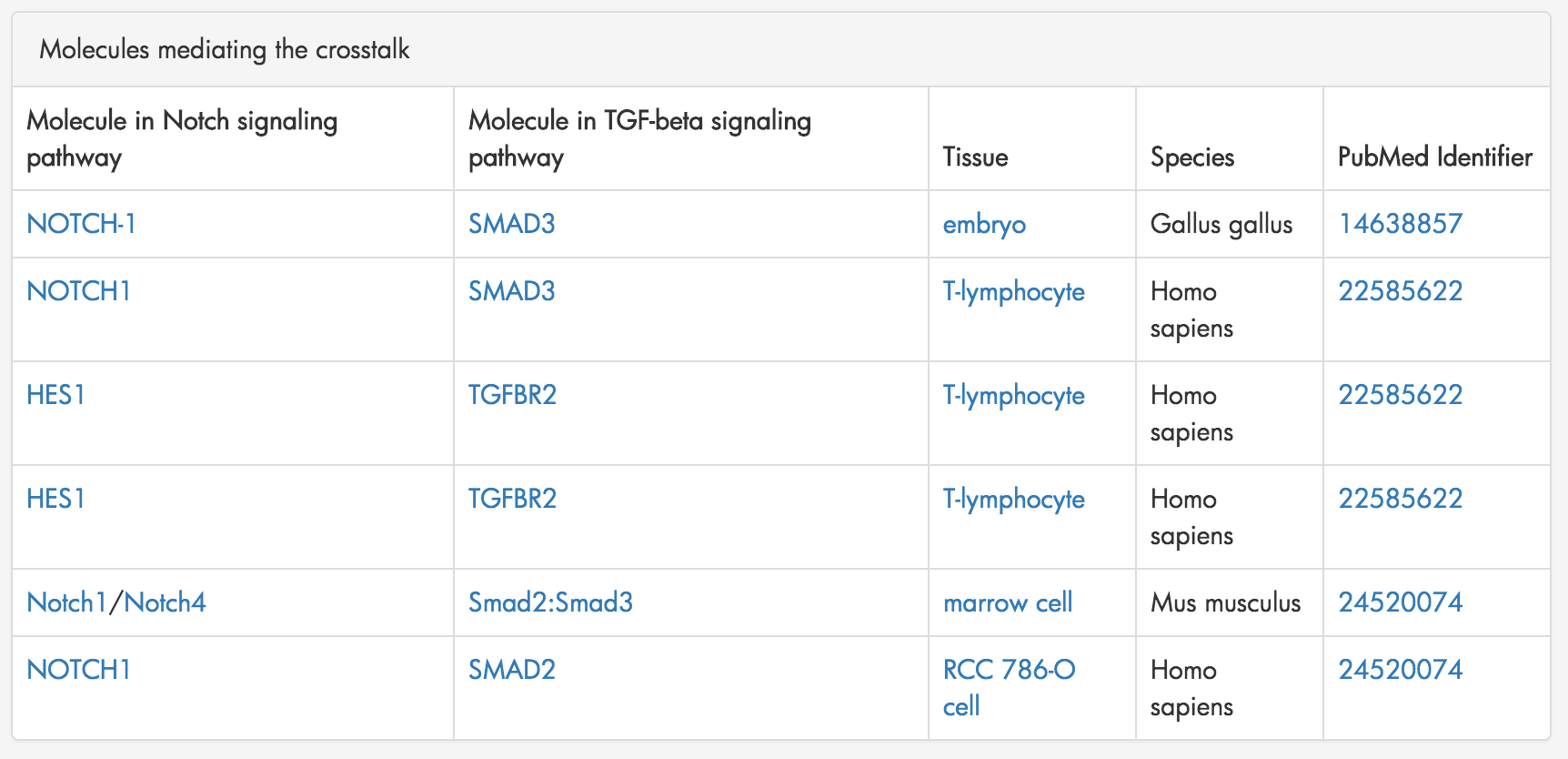
Figure 7: A table summarizing the molecular interaction that mediate crosstalk from the Notch Signaling pathway to the TGF-beta Signaling pathway.
XTalkDB displays a table summarizing all the molecules mediating the crosstalk between the two pathways. Figure 7. This table includes the molecules from each pathway, the species and tissue in which the interaction was documented and the PubMed ID corresponding to the interaction.
Network View
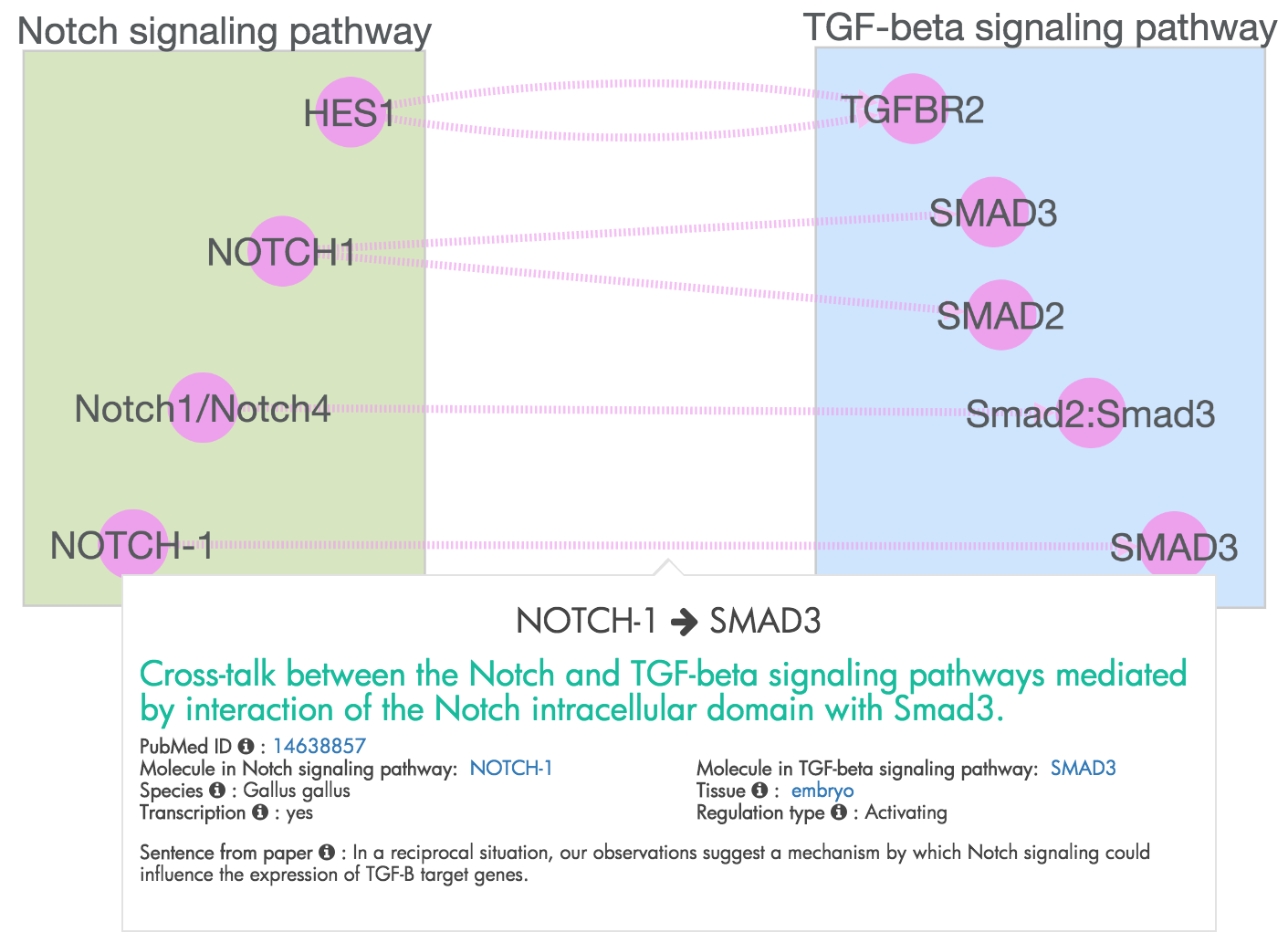
Figure 8: The network of molecular interactions mediating crosstalk from the Notch Signaling pathway to the TGF-beta Signaling pathway.
XTalkDB documents from each publication the molecules (when available) that mediate the crosstalk between two pathways. We provide these molecules as an interactive network of interactions. Figure 8. In these networks, the nodes represent the mediating molecules and the directed edge represents a relationship between molecules. The molecules are grouped by the pathway in which they participate. The user may click on an edge to view the source publication for the mediating molecules.
Downloads
The download page allows the user to download the entire database in three formats: JSON, CSV and TSV. A user may also download pathway-specific crosstalk using the download links next to each pathway name.
The downloadable file is a complete snapshot of data recorded during the curation procedure. It includes all curated publications with/without evidence for crosstalk for a given pathway pair. For each (pathway A, pathway B, PubMed ID) triplet we recorded the following information from the publication: whether the crosstalk was transcriptional, regulation type, molecule A, molecule B, species, tissue, condition, and the sentence supporting the crosstalk between the two pathways. In addition, if either molecule A or molecule B was a protein, we recorded its UniProt identi er as another attribute and "UniProt" as the source database.
| Attribute Name | Description |
|---|---|
| Pathway A | The name of the upstream (first) pathway in a pair of crosstalking pathways. |
| Pathway B | The name of the downstream (second) pathway in a pair of crosstalking pathways. |
| Pubmed Query | The string used as a structured query in PubMed that returned the recorded PMID as a result. |
| PMID | The PubMed identifier for the reported publication. |
| Crosstalk | yes, if Pathway A elicits a downstream transcriptional response in Pathway B. |
| Transcriptional | yes, if the crosstalk is transcriptional. |
| Regulation type | The downstream effect on Pathway B. This attribute can take one of the following two values:
|
| Molecule A* | The molecule in Pathway A responsible for mediating crosstalk to Pathway B. |
| Molecule A Identifier | Unique identifier for Molecule A in the namespace recorded in "Molecule A Source", e.g., the UniProt ID of a protein. |
| Molecule A Source | The name of database that the value in "Molecule A Identifier" comes from, e.g., "UniProt" if the molecule is a protein. |
| Molecule B* | The molecule in Pathway B responsible for mediating crosstalk from Pathway A. |
| Molecule B Identifier | Unique identifier for Molecule B in the namespace recorded in "Molecule B Source", e.g., the UniProt ID of a protein. |
| Molecule B Source | The name of database that the value in "Molecule B Identifier" comes from, e.g., "UniProt" if the molecule is a protein. |
| Species | The name of the species in which the crosstalk was observed. |
| Tissue | The name of the tissue or cell line in which the crosstalk was observed. |
| BTO ID | The BRENDA Tissue Ontology (BTO) Identifier of the tissue or cell line in which the crosstalk was observed. |
| Condition | Notes on the experimental condition in the publication. |
| Sentence from paper | The sentence in the publication supporting the crosstalk. We record a sentence only if it states that Pathway A increases or decreases Pathway B signaling. The sentence may also include information about the proteins or genes responsible for mediating the crosstalk. |
| Misleading evidence for crosstalk | A sentence in the paper that appears to support evidence for crosstalk when the study does not conclude there is crosstalk. |
| Additional notes | A curator's notes that may provide rationale for the values recorded for the attributes. |
*This attribute may represent either an individual molecule or several molecules. We use the following syntax for this attribute.
- colon (:): The molecules participate in the complex, e.g., SMAD3:SMAD4 in the case of crosstalk from the TGF-beta signaling pathway to the Hippo signaling pathway (the complex consisting of SMAD3 and SMAD4 mediates this crosstalk).
- slash (/): Either of the molecules can mediate the crosstalk, e.g., YAP1/WWTR1 for the same pair of pathways (YAP1 or WWTR1 can mediate the crosstalk).
- comma (,): All the molecules are required for the crosstalk but they do not form a complex, e.g., TSC2,RPTOR for the crosstalk from the MAPK signaling pathway to the mTOR signaling pathway (both TSC2 and RPTOR act as mediators).
- brackets ([]): If we cannot identify the specific molecule, we record all molecules in the family, e.g., [TEAD1/TEAD2/TEAD3/TEAD4]. In this case, the publication only listed the protein TEAD as mediating the crosstalk (from the Hippo signaling pathway to the Wnt signaling pathway).
Curation
Criteria for selecting pathways
We selected pathway names from the KEGG database since it has one of the most comprehensive collection of pathways. KEGG Pathways are divided into seven categories: Metabolism, Genetic Information Processing, Environmental Information Processing, Cellular Processes, Organismal Systems, Human Diseases, and Drug Development. We selected the following categories related to intracellular signaling: Environmental Information Processing (Signal transduction and Signaling molecules and interaction), Cellular Processes (Cell growth and death and Cell community), and Organismal Systems (Immune system and Endocrine system). Since the definition of crosstalk relies on the transduction of a signal from one pathway to another, we limited ourselves to those pathways that have at least one receptor and one transcription factor. See the statistics page for the list of pathways.
Procedure for Curating XTalkDB
We manually curated the literature for crosstalk between all directed pairs of pathways. For each pathway pair, we recorded whether any publications documented an experiment that demonstrated crosstalk or whether we found no such publications. We set up a detailed procedure to ensure high curation quality in XTALKDB. For every pathway pair we performed a systematic search in PubMed using structured queries to identify literature that may support their crosstalk. We searched the resulting publications to see if there was any literature support for crosstalk in either direction. We recorded 20 attributes for each pair of pathways per publication.
Our curation procedure consisted of the following steps:
- Select two pathways to be queried (referred to as “A” and “B”). For every such selection, there are two ordered pairs: (A, B) and (B, A). Note that the crosstalk from A → B is different from crosstalk from B → A.
- We searched PubMed for publications that may provide evidence for crosstalk between two pathways using a structured query. We formatted these queries as
- Structured:
- “A” AND “B” AND “crosstalk”
- “A” AND “B” AND “pathway”
- Occasionally, we modified these queries by using synonyms for pathway names or by including proteins within the pathways. For example, we used the query 'GLP-1 Adipocytokine Signaling' instead of 'Insulin Signaling AND Adipocytokine AND Pathway'.The downloadable files provided on the XTalkDB website specify the precise queries for each pathway pair.
- Structured:
- If none of the queries returned any results, we recorded each of the pathway pairs with "NO_RESULTS_FOR_PUBMED_QUERY" as a dummy PubMed Id.
- We processed the PubMed search results in order of relevance.
- If there were at most 10 publications returned, we read each abstract. Otherwise, we selected paper titles that we judged were likely to report crosstalk.
- Note that some publications may support crosstalk from pathway A to pathway B and other papers in the other direction. It is also possible for a publication to support crosstalk in both direction. We describe how we curated a paper for evidence of crosstalk from pathway A to pathway B. We used an analogous procedure for crosstalk in the other direction. In effect, we evaluated each publication twice.
- We searched the abstract for a sentence that supported crosstalk. We recorded that there was no crosstalk if the abstract had enough evidence to support this conclusion. For example, an abstract may have mentioned both pathways (among others) but did not mention them together, or the paper was included in the results because it reported intercellular crosstalk.
- If we found a sentence in the abstract that supports crosstalk or there was not enough information in the abstract to conclude that there is no crosstalk, we then searched the full text of the paper (when available) for sentences that contained the names of one or both pathways. We then read each of these sentences within its context as well as the Discussion or Conclusion section of the paper to judge their support of crosstalk.
- We also examined the publication for sentences that provided further information on crosstalk: the molecules that mediate the crosstalk, whether pathway A activates or inhibits pathway B, and the species and tissue type in which the experiment reported in the publication was performed.
- For each pair of pathways shown to crosstalk in a publication, we recorded the following information in the database: whether the crosstalk was transcriptional, the regulation type (activating or inhibiting), molecular component A, molecular component B, species, tissue, condition, and the sentence supporting the crosstalk between the two pathways. Whenever possible, we mapped the molecules and tissues to a common namespace. See the table of attributes for a description of all the features recorded in XTalkDB.
- If the publication did not support crosstalk, we recorded that this publication had no evidence for crosstalk from pathway A to pathway B.
Additional notes on the curation procedure
- If the full text of a paper was not available, we recorded as much information (on support for crosstalk or lack thereof) from the abstract.
- On occasion, a publication returned by our search query was a review article. In such a case, we recorded the information documenting crosstalk available in the review article instead of the original source article that reported the crosstalk. We opted for this strategy since we wanted to document which paper and sentence led to the entry in XTalkDB.
- Through this curation procedure, we recorded (pathway A, pathway B, PubMed ID) triples where the publication did not provide support for crosstalk from pathway A to pathway B. These triples are part of the file downloads on the XTalkDB website. However, the search interface does not include them in the results.
- It was not possible to confirm that a pathway pair does not crosstalk. What we could conclude from this process was that none of the publications that we have curated contained evidence for crosstalk between that pair of pathways.
- Publications that we curated typically monitored the activity [TMM 1: Is there a different word to use here? Otherwise, activity/activation appears too many times in this note.] of a pathway by measuring the expression of a target (reporter) gene or the activity of a representative protein. We recorded information on activation and inhibition between pathway pairs as follows: if a publication reported that the stimulation of pathway A activated (respectively, inhibited) pathway B, then we considered pathway A to activate (respectively, inhibit) pathway B. [TMM 2: Need to work on this sentence.]
- The molecules that mediate the crosstalk between a pair of pathways may not directly interact. Hence, these molecules do not always provide information on the mechanism underlying the crosstalk.
Curation Rules
We followed the following rules while mapping the molecules to their corresponding nomenclature:
- If the mediating protein is not explicitly made clear in the text, check the materials and methods section of the manuscript for more specific information (e.g., which antibodies were used, which primers, etc.)
- If an assumption is made about which protein (e.g., VEGFa for all VEGF), then make a note in the curators notes.
- If the organism/species does not contain the identified protein (for example, transgenic species), use the UniProt id in the original organism, and add an explanation in the "Curator Notes", e.g., "Using the UniProt id for ERa in Homo sapiens, since ERa was ectopically expressed in Drosophila melanogaster, but UniProt does not contain an id for this protein in this organism.
- If the identified protein in a given species does not have a reviewed UniProt ID. For these proteins, we include the first unreviewed UniProt ID listed by UniProt.
- If the identified molecule is not a protein (e.g., amino acid or miRNAs), we record the name and type of the molecule. Note: We will be mapping these molecules to their corresponding namespaces in future.